Essential idea: Chromosomes carry genes in a linear sequence that is shared by members of a species.
The asian rice (Oryza sativa) genome can be seen illustrated above. Rice possesses up 63,000 genes divided up between 12 chromosomes.
Above to the right is a map of the first chromosome showing all the gene loci present on it. Although different varieties (estimated 40,000 worldwide) will possess different alleles for genes, all individuals will share the same twelve chromosomes and the alleles of each variety will occur at the same position on same chromosome, i.e. at the same gene loci.
Above to the right is a map of the first chromosome showing all the gene loci present on it. Although different varieties (estimated 40,000 worldwide) will possess different alleles for genes, all individuals will share the same twelve chromosomes and the alleles of each variety will occur at the same position on same chromosome, i.e. at the same gene loci.
Understandings, applications and skills:
| 3.2.U1 | Prokaryotes have one chromosome consisting of a circular DNA molecule. |
| 3.2.U2 | Some prokaryotes also have plasmids but eukaryotes do not. |
| 3.2.U3 | Eukaryote chromosomes are linear DNA molecules associated with histone proteins. |
| 3.2.U4 | In a eukaryote species there are different chromosomes that carry different genes. |
| 3.2.U5 | Homologous chromosomes carry the same sequence of genes but not necessarily the same alleles of those genes. |
| 3.2.U6 | Diploid nuclei have pairs of homologous chromosomes. |
| 3.2.U7 | Haploid nuclei have one chromosome of each pair. [The two DNA molecules formed by DNA replication prior to cell division are considered to be sister chromatids until the splitting of the centromere at the start of anaphase. After this, they are individual chromosomes.] |
| 3.2.U8 | The number of chromosomes is a characteristic feature of members of a species. |
| 3.2.U9 | A karyogram shows the chromosomes of an organism in homologous pairs of decreasing length. [The terms karyotype and karyogram have different meanings. Karyotype is a property of a cell—the number and type of chromosomes present in the nucleus, not a photograph or diagram of them.] |
| 3.2.U10 | Sex is determined by sex chromosomes and autosomes are chromosomes that do not determine sex. |
| 3.2.A1 | Cairns’ technique for measuring the length of DNA molecules by autoradiography. |
| 3.2.A2 | Comparison of genome size in T2 phage,Escherichia coli, Drosophila melanogaster, Homo sapiens and Paris japonica. [Genome size is the total length of DNA in an organism. The examples of genome and chromosome number have been selected to allow points of interest to be raised.] |
| 3.2.A3 | Comparison of diploid chromosome numbers of Homo sapiens, Pan troglodytes, Canis familiaris, Oryza sativa, Parascaris equorum. |
| 3.2.A4 | Use of karyograms to deduce sex and diagnose Down syndrome in humans. |
| 3.2.S1 | Use of databases to identify the locus of a human gene and its polypeptide product. |
[Text in square brackets indicates guidance notes]
Presentation and Notes
The presentation is designed to help your understanding. The notes outline is intended to be used as a framework for the development of student notes to aid revision.
|
|
Nature of science:
Developments in research follow improvements in techniques—autoradiography was used to establish the length of DNA molecules in chromosomes. (1.8) [Linked to 3.2.A1]
International-mindedness:
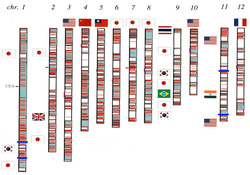
Sequencing of the rice genome involved cooperation between biologists in 10 countries.
The acknowledgement list below, and the genome map to the right (source: http://www.patentlens.net/daisy/RiceGenome/2738.html) illustrate the size and international nature of teams that scientists work in. Teams are often loose collaborations between scientists working in different institutions.
Acknowledgments
Physical Maps and Sequencing: Rice Genome Research Program (RGP)Takashi Matsumoto1, Jianzhong Wu1, Hiroyuki Kanamori1, Yuichi Katayose1, Masaki Fujisawa1, Nobukazu Namiki1, Hiroshi Mizuno1, Kimiko Yamamoto1, Baltazar A. Antonio1, Tomoya Baba1, Katsumi Sakata1, Yoshiaki Nagamura1, Hiroyoshi Aoki1, Koji Arikawa1, Kohei Arita1, Takahito Bito1, Yoshino Chiden1, Nahoko Fujitsuka1, Rie Fukunaka1, Masao Hamada1, Chizuko Harada1, Akiko Hayashi1, Saori Hijishita1, Mikiko Honda1, Satomi Hosokawa1, Yoko Ichikawa1, Atsuko Idonuma1, Masumi Iijima1, Michiko Ikeda1, Maiko Ikeno1, Kazue Ito1, Sachie Ito1, Tomoko Ito1, Yuichi Ito1, Yukiyo Ito1, Aki Iwabuchi1, Kozue Kamiya1, Wataru Karasawa1, Kanako Kurita1, Satoshi Katagiri1, Ari Kikuta1, Harumi Kobayashi1, Noriko Kobayashi1, Kayo Machita1, Tomoko Maehara1, Masatoshi Masukawa1, Tatsumi Mizubayashi1, Yoshiyuki Mukai1, Hideki Nagasaki1, Yuko Nagata1, Shinji Naito1, Marina Nakashima1, Yuko Nakama1, Yumi Nakamichi1, Mari Nakamura1, Ayano Meguro1, Manami Negishi1, Isamu Ohta1, Tomoya Ohta1, Masako Okamoto1, Nozomi Ono1, Shoko Saji1, Miyuki Sakaguchi1, Kumiko Sakai1, Michie Shibata1, Takanori Shimokawa1, Jianyu Song1, Yuka Takazaki1, Kimihiro Terasawa1, Mika Tsugane1, Kumiko Tsuji1, Shigenori Ueda1, Kazunori Waki1, Harumi Yamagata1, Mayu Yamamoto1, Shinichi Yamamoto1, Hiroko Yamane1, Shoji Yoshiki1, Rie Yoshihara1, Kazuko Yukawa1, Huisun Zhong1, Masahiro Yano1, Takuji Sasaki, (Principal Investigator)1; The Institute for Genomic Research (TIGR) Qiaoping Yuan2, Shu Ouyang2, Jia Liu2, Kristine M. Jones2, Kristen Gansberger2, Kelly Moffat2, Jessica Hill2, Jayati Bera2, Douglas Fadrosh2, Shaohua Jin2, Shivani Johri2, Mary Kim2, Larry Overton2, Matthew Reardon2, Tamara Tsitrin2, Hue Vuong2, Bruce Weaver2, Anne Ciecko2, Luke Tallon2, Jacqueline Jackson2, Grace Pai2, Susan Van Aken2, Terry Utterback2, Steve Reidmuller2, Tamara Feldblyum2, Joseph Hsiao2, Victoria Zismann2, Stacey Iobst2, Aymeric R. de Vazeille2, C. Robin Buell, (Principal Investigator)2; National Center for Gene Research Chinese Academy of Sciences (NCGR) Kai Ying3, Ying Li3, Tingting Lu3, Yuchen Huang3, Qiang Zhao3, Qi Feng3, Lei Zhang3, Jingjie Zhu3, Qijun Weng3, Jie Mu3, Yiqi Lu3, Danlin Fan3, Yilei Liu3, Jianping Guan3, Yujun Zhang3, Shuliang Yu3, Xiaohui Liu3, Yu Zhang3, Guofan Hong3, Bin Han, (Principal Investigator)3, Genoscope, Nathalie Choisne4, Nadia Demange4, Gisela Orjeda4, Sylvie Samain4, Laurence Cattolico4, Eric Pelletier4, Arnaud Couloux4, Beatrice Segurens4, Patrick Wincker4, Angelique D'Hont5, Claude Scarpelli4, Jean Weissenbach4, Marcel Salanoubat4, Francis Quetier, (Principal Investigator)4; Arizona Genomics Institute (AGI) and Arizona Genomics Computational Laboratory (AGCol) Yeisoo Yu6, Hye Ran Kim6, Teri Rambo6, Jennifer Currie6, Kristi Collura6, Meizhong Luo6, Tae-Jin Yang6, Jetty S. S. Ammiraju6, Friedrich Engler6, Carol Soderlund6, Rod A. Wing, (Principal Investigator)6; Cold Spring Harbor Laboratory (CSHL) Lance E. Palmer7, Melissa de la Bastide7, Lori Spiegel7, Lidia Nascimento7, Theresa Zutavern7, Andrew O'Shaughnessy7, Sujit Dike7, Neilay Dedhia7, Raymond Preston7, Vivekanand Balija7, W. Richard McCombie, (Principal Investigator)7; Academia Sinica Plant Genome Center (ASPGC) Teh-Yuan Chow8, Hong-Hwa Chen9, Mei-Chu Chung8, Ching-San Chen8, Jei-Fu Shaw8, Hong-Pang Wu8, Kwang-Jen Hsiao10, Ya-Ting Chao8, Mu-kuei Chu8, Chia-Hsiung Cheng8, Ai-Ling Hour8, Pei-Fang Lee8, Shu-Jen Lin8, Yao-Cheng Lin8, John-Yu Liou8, Shu-Mei Liu8, Yue-Ie Hsing, (Principal Investigator)8; Indian Initiative for Rice Genome Sequencing (IIRGS), University of Delhi South Campus (UDSC) S. Raghuvanshi11, A. Mohanty11, A. K. Bharti11,13, A. Gaur11, V. Gupta11, D. Kumar11, V. Ravi11, S. Vij11, A. Kapur11, Parul Khurana11, Paramjit Khurana11, J. P. Khurana11, A. K. Tyagi, (Principal Investigator)11; Indian Initiative for Rice Genome Sequencing (IIRGS), Indian Agricultural Research Institute (IARI) K. Gaikwad12, A. Singh12, V. Dalal12, S. Srivastava12, A. Dixit12, A. K. Pal12, I. A. Ghazi12, M. Yadav12, A. Pandit12, A. Bhargava12, K. Sureshbabu12, K. Batra12, T. R. Sharma12, T. Mohapatra12, N. K. Singh, (Principal Investigator)12; Plant Genome Initiative at Rutgers (PGIR) Joachim Messing, (Principal Investigator)13, Amy Bronzino Nelson13, Galina Fuks13, Steve Kavchok13, Gladys Keizer13, Eric Linton Victor Llaca13, Rentao Song13, Bahattin Tanyolac13, Steve Young13; Korea Rice Genome Research Program (KRGRP) Kim Ho-Il14, Jang Ho Hahn, (Principal Investigator)14; National Center for Genetic Engineering and Biotechnology (BIOTEC) G. Sangsakoo15, A. Vanavichit, (Principal Investigator)15; Brazilian Rice Genome Initiative (BRIGI) Luiz Anderson Teixeira de Mattos16, Paulo Dejalma Zimmer16, Gaspar Malone16, Odir Dellagostin16, Antonio Costa de Oliveira, (Principal Investigator)16; John Innes Centre (JIC) Michael Bevan17, Ian Bancroft17; Washington University School of Medicine Genome Sequencing Center Pat Minx18, Holly Cordum18, Richard Wilson18; University of Wisconsin-Madison Zhukuan Cheng19, Weiwei Jin19, Jiming Jiang19, Sally Ann Leong20
Annotation and Analysis: Hisakazu Iwama21, Takashi Gojobori21,22, Takeshi Itoh22,23, Yoshihito Niimura24, Yasuyuki Fujii25, Takuya Habara25, Hiroaki Sakai23,25, Yoshiharu Sato22, Greg Wilson26, Kiran Kumar27, Susan McCouch26, Nikoleta Juretic28, Douglas Hoen28, Stephen Wright29, Richard Bruskiewich30, Thomas Bureau28, Akio Miyao23, Hirohiko Hirochika23, Tomotaro Nishikawa23, Koh-ichi Kadowaki23, Masahiro Sugiura31
Coordination: Benjamin Burr32
To read more about the rice genome you can read this this article by published in Nature
The acknowledgement list below, and the genome map to the right (source: http://www.patentlens.net/daisy/RiceGenome/2738.html) illustrate the size and international nature of teams that scientists work in. Teams are often loose collaborations between scientists working in different institutions.
Acknowledgments
Physical Maps and Sequencing: Rice Genome Research Program (RGP)Takashi Matsumoto1, Jianzhong Wu1, Hiroyuki Kanamori1, Yuichi Katayose1, Masaki Fujisawa1, Nobukazu Namiki1, Hiroshi Mizuno1, Kimiko Yamamoto1, Baltazar A. Antonio1, Tomoya Baba1, Katsumi Sakata1, Yoshiaki Nagamura1, Hiroyoshi Aoki1, Koji Arikawa1, Kohei Arita1, Takahito Bito1, Yoshino Chiden1, Nahoko Fujitsuka1, Rie Fukunaka1, Masao Hamada1, Chizuko Harada1, Akiko Hayashi1, Saori Hijishita1, Mikiko Honda1, Satomi Hosokawa1, Yoko Ichikawa1, Atsuko Idonuma1, Masumi Iijima1, Michiko Ikeda1, Maiko Ikeno1, Kazue Ito1, Sachie Ito1, Tomoko Ito1, Yuichi Ito1, Yukiyo Ito1, Aki Iwabuchi1, Kozue Kamiya1, Wataru Karasawa1, Kanako Kurita1, Satoshi Katagiri1, Ari Kikuta1, Harumi Kobayashi1, Noriko Kobayashi1, Kayo Machita1, Tomoko Maehara1, Masatoshi Masukawa1, Tatsumi Mizubayashi1, Yoshiyuki Mukai1, Hideki Nagasaki1, Yuko Nagata1, Shinji Naito1, Marina Nakashima1, Yuko Nakama1, Yumi Nakamichi1, Mari Nakamura1, Ayano Meguro1, Manami Negishi1, Isamu Ohta1, Tomoya Ohta1, Masako Okamoto1, Nozomi Ono1, Shoko Saji1, Miyuki Sakaguchi1, Kumiko Sakai1, Michie Shibata1, Takanori Shimokawa1, Jianyu Song1, Yuka Takazaki1, Kimihiro Terasawa1, Mika Tsugane1, Kumiko Tsuji1, Shigenori Ueda1, Kazunori Waki1, Harumi Yamagata1, Mayu Yamamoto1, Shinichi Yamamoto1, Hiroko Yamane1, Shoji Yoshiki1, Rie Yoshihara1, Kazuko Yukawa1, Huisun Zhong1, Masahiro Yano1, Takuji Sasaki, (Principal Investigator)1; The Institute for Genomic Research (TIGR) Qiaoping Yuan2, Shu Ouyang2, Jia Liu2, Kristine M. Jones2, Kristen Gansberger2, Kelly Moffat2, Jessica Hill2, Jayati Bera2, Douglas Fadrosh2, Shaohua Jin2, Shivani Johri2, Mary Kim2, Larry Overton2, Matthew Reardon2, Tamara Tsitrin2, Hue Vuong2, Bruce Weaver2, Anne Ciecko2, Luke Tallon2, Jacqueline Jackson2, Grace Pai2, Susan Van Aken2, Terry Utterback2, Steve Reidmuller2, Tamara Feldblyum2, Joseph Hsiao2, Victoria Zismann2, Stacey Iobst2, Aymeric R. de Vazeille2, C. Robin Buell, (Principal Investigator)2; National Center for Gene Research Chinese Academy of Sciences (NCGR) Kai Ying3, Ying Li3, Tingting Lu3, Yuchen Huang3, Qiang Zhao3, Qi Feng3, Lei Zhang3, Jingjie Zhu3, Qijun Weng3, Jie Mu3, Yiqi Lu3, Danlin Fan3, Yilei Liu3, Jianping Guan3, Yujun Zhang3, Shuliang Yu3, Xiaohui Liu3, Yu Zhang3, Guofan Hong3, Bin Han, (Principal Investigator)3, Genoscope, Nathalie Choisne4, Nadia Demange4, Gisela Orjeda4, Sylvie Samain4, Laurence Cattolico4, Eric Pelletier4, Arnaud Couloux4, Beatrice Segurens4, Patrick Wincker4, Angelique D'Hont5, Claude Scarpelli4, Jean Weissenbach4, Marcel Salanoubat4, Francis Quetier, (Principal Investigator)4; Arizona Genomics Institute (AGI) and Arizona Genomics Computational Laboratory (AGCol) Yeisoo Yu6, Hye Ran Kim6, Teri Rambo6, Jennifer Currie6, Kristi Collura6, Meizhong Luo6, Tae-Jin Yang6, Jetty S. S. Ammiraju6, Friedrich Engler6, Carol Soderlund6, Rod A. Wing, (Principal Investigator)6; Cold Spring Harbor Laboratory (CSHL) Lance E. Palmer7, Melissa de la Bastide7, Lori Spiegel7, Lidia Nascimento7, Theresa Zutavern7, Andrew O'Shaughnessy7, Sujit Dike7, Neilay Dedhia7, Raymond Preston7, Vivekanand Balija7, W. Richard McCombie, (Principal Investigator)7; Academia Sinica Plant Genome Center (ASPGC) Teh-Yuan Chow8, Hong-Hwa Chen9, Mei-Chu Chung8, Ching-San Chen8, Jei-Fu Shaw8, Hong-Pang Wu8, Kwang-Jen Hsiao10, Ya-Ting Chao8, Mu-kuei Chu8, Chia-Hsiung Cheng8, Ai-Ling Hour8, Pei-Fang Lee8, Shu-Jen Lin8, Yao-Cheng Lin8, John-Yu Liou8, Shu-Mei Liu8, Yue-Ie Hsing, (Principal Investigator)8; Indian Initiative for Rice Genome Sequencing (IIRGS), University of Delhi South Campus (UDSC) S. Raghuvanshi11, A. Mohanty11, A. K. Bharti11,13, A. Gaur11, V. Gupta11, D. Kumar11, V. Ravi11, S. Vij11, A. Kapur11, Parul Khurana11, Paramjit Khurana11, J. P. Khurana11, A. K. Tyagi, (Principal Investigator)11; Indian Initiative for Rice Genome Sequencing (IIRGS), Indian Agricultural Research Institute (IARI) K. Gaikwad12, A. Singh12, V. Dalal12, S. Srivastava12, A. Dixit12, A. K. Pal12, I. A. Ghazi12, M. Yadav12, A. Pandit12, A. Bhargava12, K. Sureshbabu12, K. Batra12, T. R. Sharma12, T. Mohapatra12, N. K. Singh, (Principal Investigator)12; Plant Genome Initiative at Rutgers (PGIR) Joachim Messing, (Principal Investigator)13, Amy Bronzino Nelson13, Galina Fuks13, Steve Kavchok13, Gladys Keizer13, Eric Linton Victor Llaca13, Rentao Song13, Bahattin Tanyolac13, Steve Young13; Korea Rice Genome Research Program (KRGRP) Kim Ho-Il14, Jang Ho Hahn, (Principal Investigator)14; National Center for Genetic Engineering and Biotechnology (BIOTEC) G. Sangsakoo15, A. Vanavichit, (Principal Investigator)15; Brazilian Rice Genome Initiative (BRIGI) Luiz Anderson Teixeira de Mattos16, Paulo Dejalma Zimmer16, Gaspar Malone16, Odir Dellagostin16, Antonio Costa de Oliveira, (Principal Investigator)16; John Innes Centre (JIC) Michael Bevan17, Ian Bancroft17; Washington University School of Medicine Genome Sequencing Center Pat Minx18, Holly Cordum18, Richard Wilson18; University of Wisconsin-Madison Zhukuan Cheng19, Weiwei Jin19, Jiming Jiang19, Sally Ann Leong20
Annotation and Analysis: Hisakazu Iwama21, Takashi Gojobori21,22, Takeshi Itoh22,23, Yoshihito Niimura24, Yasuyuki Fujii25, Takuya Habara25, Hiroaki Sakai23,25, Yoshiharu Sato22, Greg Wilson26, Kiran Kumar27, Susan McCouch26, Nikoleta Juretic28, Douglas Hoen28, Stephen Wright29, Richard Bruskiewich30, Thomas Bureau28, Akio Miyao23, Hirohiko Hirochika23, Tomotaro Nishikawa23, Koh-ichi Kadowaki23, Masahiro Sugiura31
Coordination: Benjamin Burr32
To read more about the rice genome you can read this this article by published in Nature

